Environment cues¶
Querying the environment for cues that affect Front growth is an important component of a simulation. During a manage_front call, the following data can be obtained:
Location of all nearby
Frontof the sameNeuron:get_frontsmethod, useful to model self-repulsion between dendrites.Location of all nearby
Frontof otherNeuron:get_frontsmethod, useful to model attraction or repulsion by other neuron dendrites or axons.Location or local concentration of
Substrate:get_substratesmethod, simulates chemical attraction independent ofFrontstructures.a CollisionError: contains information about the colliding
Front, see Preventing and dealing with collisions.
get_fronts method¶
get_fronts is a Front method, usually called by self. It returns a list of tuples: (Front, distance), where distance is the shortest distance between self (or the calling front) and Front. By default, this list will be sorted with nearest fronts first and only fronts within a range of 100 µm will be searched. With optional parameter returnID=True (ID, distance) tuples will be returned instead.
The main parameter for get_fronts is the optional what, a string that determines which fronts will be returned:
‘self’: get fronts belonging to same neuron as self, excluding all up to second order ancestors and descendents.
‘self+’: get all fronts within max_distance belonging to self.
‘name’: get fronts belonging to neurons with a name (wildcard), not including same neuron as self.
‘other’: get fronts that do not belong to self (default).
‘type’: get fronts belonging to a type of neuron specified in name, not including same neuron as self.
See simulator module for complete documentation of the get_fronts method.
A simple example of self-repulsion, only by nearby fronts within 20 µm:
def manage_front(self,constellation):
others = self.get_fronts(constellation,what="self",max_distance=20.)
if others: # nearby fronts of same neuron found, excluding parent and children
nearest = goals[0][0] # get nearest front of same neuron
dir_to_repel = nearest.mid() - self.end # compute direction to nearest front
else: # no repelling fronts found
dir_to_repel = Point(0.,0.,0.) # no repulsion
new_pos = self.end + self.unit_heading_sample(width=20.) * 5. - dir_to_repel.norm() * 2.
A realistic self-repulsion model has of course to deal with all nearby fronts, not just the nearest one, which may not be trivial.
An example of attraction to a named neuron, from the Environment notebook:
def manage_front(self,constellation):
goals = self.get_fronts(constellation,what="name",name="attract_neuron")
# use first of the list
if goals:
goal_front = goals[0][0] # get nearest atract_neuron front
dir_to_goal = goal_front.end - self.end # compute direction to nearest front
else: # deal with absence of attractor
dir_to_goal = self.unit_heading_sample(width=10.)
new_pos = self.end + dir_to_goal.norm() * 5.0
The run-time of get_fronts scales with the number of neurons and fronts in the simulation and may become quite slow for very large simulations. Therefore, an alternative faster search method is implemented if only nearby fronts are desired, this method will be automatically used if optional parameter max_distance <= Admin_agent.grid_step.
Inside versus outside of a front¶
The code examples above computed a direction to one of the Front coordinates, which is inside the target front. This is fine for repulsion, but if the goal is to grow close to the target front, for example to make a synapse, points on the surface of the front are more relevant. This can be obtained with the surface_point_to method that returns a point on the surface of the calling front in the direction of a given other point:
def manage_front(self,constellation):
# find a front to grow toward
goals = self.get_fronts(constellation,what="name",name="axons")
if goals:
nearest = goals[0][0] # get nearest axon front
goal = nearest.surface_point_to(self) # point on surface of nearest towards front calling manage_front
direction = goal - self.end # direction to goal on nearest
distance = direction.length() # distance to goal
By default surface_point_to returns a point halfway along the length of a cylindrical front (for a sphere it is the nearest surface point). This can be changed either to a random location (optional parameter mid=False) or to a specific location along the length (e.g. for first third, optional parameter pos=0.33).
Finally, it is also possible to request a point some distance away from the front surface using the offset optional parameter. This may be helpful to prevent a collision with the target nearest:
def manage_front(self,constellation):
...
goal = nearest.surface_point_to(self,offset=0.2)
try:
new_front = self.add_child(constellation,goal) # make a new front ending close to nearest
...
Chemical cue using Substrate¶
Substrate implements modeling of chemical cues that can be placed anywhere in the simulation volume. They can be found with the get_substrates method, always based on the name of the Substrate:
def manage_front(self,constellation):
...
substrates = self.get_substrates(constellation,"attractor")
if substrates:
closest = substrates[0][0]
cdistance = substrates[0][1]
else:
...
Similar to get_fronts, this method returns a list of (Front, distance) or (ID, distance) tuples.
Substrate can be used in two different ways, both are illustrated in the Environment notebook.
The simplest is to use it as a deterministic cue and compute the direction to it:
dir_to_sub = closest.orig - self.end # compute direction to attractor
new_pos = self.end + dir_to_sub.norm() * 5.0
A bit more sophisticated is to include a dependence on distance:
if cdistance <= 2.: # go directly
new_pos = closest.orig
elif cdistance <= 5.: # approach directly in small steps
new_pos = self.end + dir_to_sub.norm() * 2.0
else: # noisy approach
new_pos = self.end + unit_sample_on_sphere() * 2.0 + dir_to_sub.norm() * 2.0
The above code assumes that get_substrates is called every cycle, a faster alternative is to store the ID as illustrated in the Environment notebook but then cdistance has to be computed every cycle.
A completely different approach to using Substrate is stochastic, this assumes that Substrate was initiated with the relevant parameters. This approach uses the diff_gradient_to method to compute a stochastic number of substrate molecules at a given Point and the direction towards the substrate at this point:
def manage_front(self,constellation):
...
substrates = self.get_substrates(constellation,"attractor")
# nmols is stochastic integer number of molecules, sdir is a Point vector towards substrate
n_mols,sdir = diff_gradient_to(self.end,substrates,constellation.cycle)
# stronger signal produces less noisy direction vector
dir_to_attractor = sdir * n_mols + rnd_dir * 1.5
new_pos = self.end + dir_to_attractor.norm() * 3.
Depending on how Substrate was initiated, the stochastic number of molecules is either computed for a continuously producing point source in infinite medium (substrate.rate > 0.) or for an instantaneous point source in infinite medium (substrate.rate = 0.). Note that these calculations make strong simplifying assumptions and may therefore not be very realistic, especially in small crowded environments or with multiple locations of the substrate. An example of the stochastic number of molecules returned at different locations by diff_gradient_to for the steady state of a continuously producing source in the upper right corner is shown in the figure:
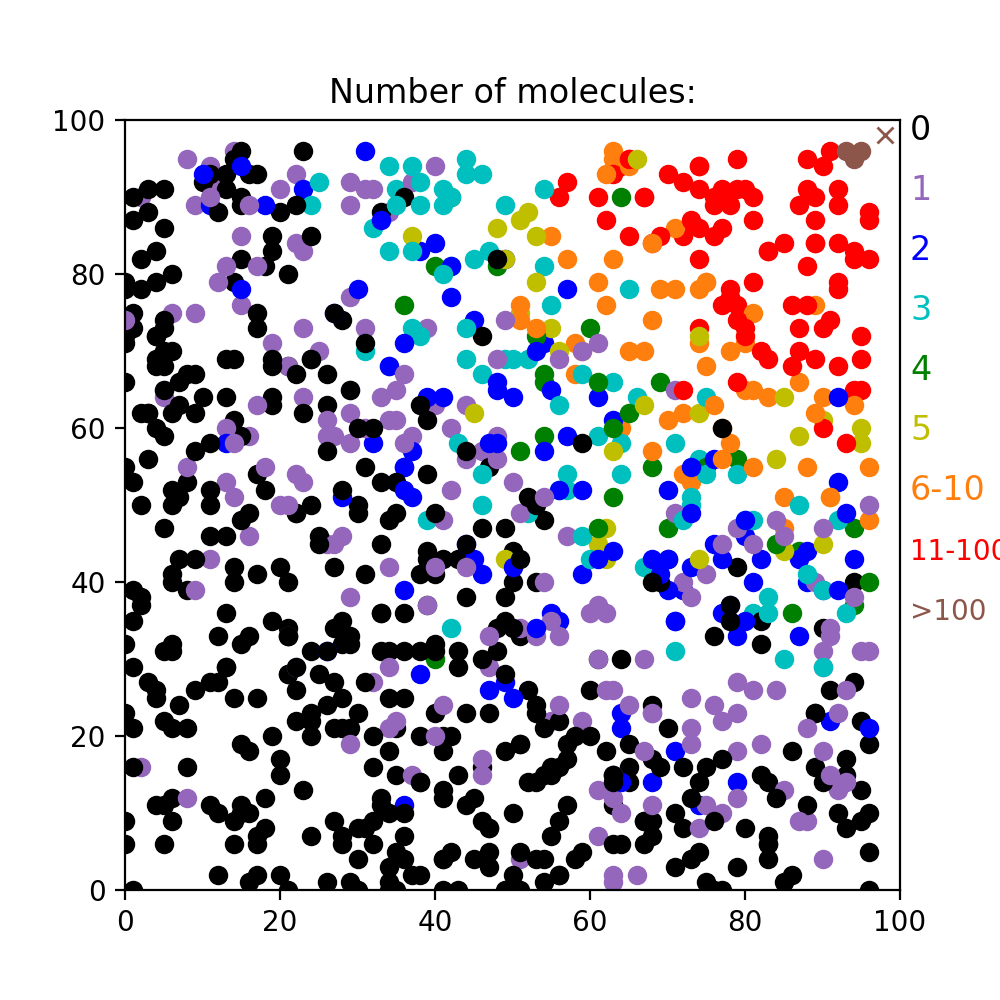
The steady state was obtained by passing -1 instead of the cycle:
n_mols,sdir = diff_gradient_to(self.end,substrates,-1)
Note that the entire substrates list is passed to diff_gradient_to. If this list contains multiple substrate sources, by default diff_gradient_to will pick the nearest one, but there is also an option to compute an average location (optional parameter what="average"). Note that diff_gradient_to always expects a list, but this can also be just a list of substrates (e.g. [Sub1,Sub2] or [Sub1]) instead of the list of tuples returned by get_substrates. The level of stochasticity can be controlled by the optional size parameter that controls the size of the sampling box.