Modeling growth¶
Growth is simulated in NeuroDevSim by creating new fronts that will become the child of the front calling self.manage_front. This tutorial section focuses on implementing specific kinds of phenomenological growth, it assumes that the reader understands the coding style introduced in Getting started.
Front extension¶
Front extension is done by the add_child method. It is the simplest way to grow a neuron. It just requires a coordinate new_pos for the new front:
def manage_front(self,constellation):
...
while count < 100:
new_dir = ... # do something to get a Point with a new direction
new_pos = self.end + new_dir # compute position of child end
# check for possible collisions
try:
new_front = self.add_child(constellation,new_pos) # make a new front
# make old front inactive: stops growing -> will not call this method again
self.disable(constellation)
return # done for this cycle
except (CollisionError, GridCompetitionError, InsideParentError, VolumeError):
count += 1
continue # pick another new_pos, no attempt to correct the error
The new_front will be a cylinder with new_front.orig == self.end and new_front.end == new_pos. By default, the new front gets the same radius, branch_name and swc_type values as self, but these can easily be changed in the add_child call:
new_front = self.add_child(constellation,new_pos,radius=2.,swc_type=2,branch_name="axon1")
This section deals with successful growth, see Preventing and dealing with collisions for more advanced handling of CollisionError.
The art of front extension is determining the new direction for growth. The easiest approach is to use the unit_heading_sample method that mimics biological growth: growth cones tend to continue along their current path, called heading, with some randomness. The unit_heading_sample returns a Point vector (relative to [0, 0, 0]) that on average falls within a cone centered around the heading of the current front self:
def manage_front(self,constellation):
...
new_dir = self.unit_heading_sample()
The figure below shows different colored directions obtained by 20 calls of unit_heading_sample for the red front.
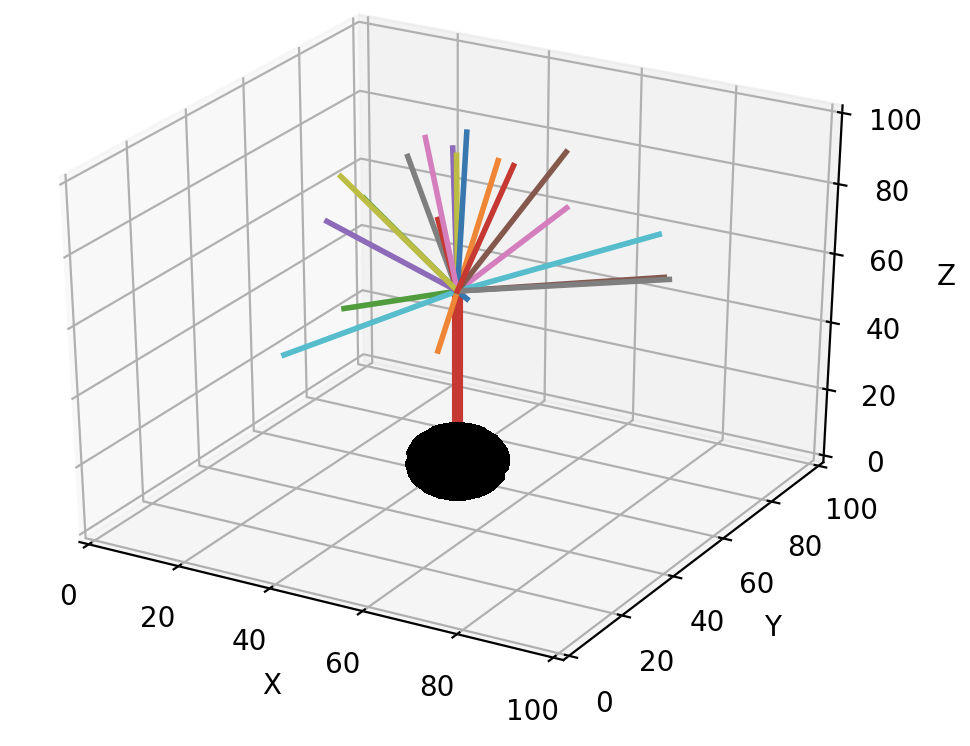
The size of the cone can be controlled with the width optional parameter, which sets the standard deviation of the Gaussian distribution around zero angle. The default value for width is 55 degrees, based on cat spinal cord motor neurons, and often a smaller cone is desirable:
def manage_front(self,constellation):
...
new_dir = self.unit_heading_sample(width=20)
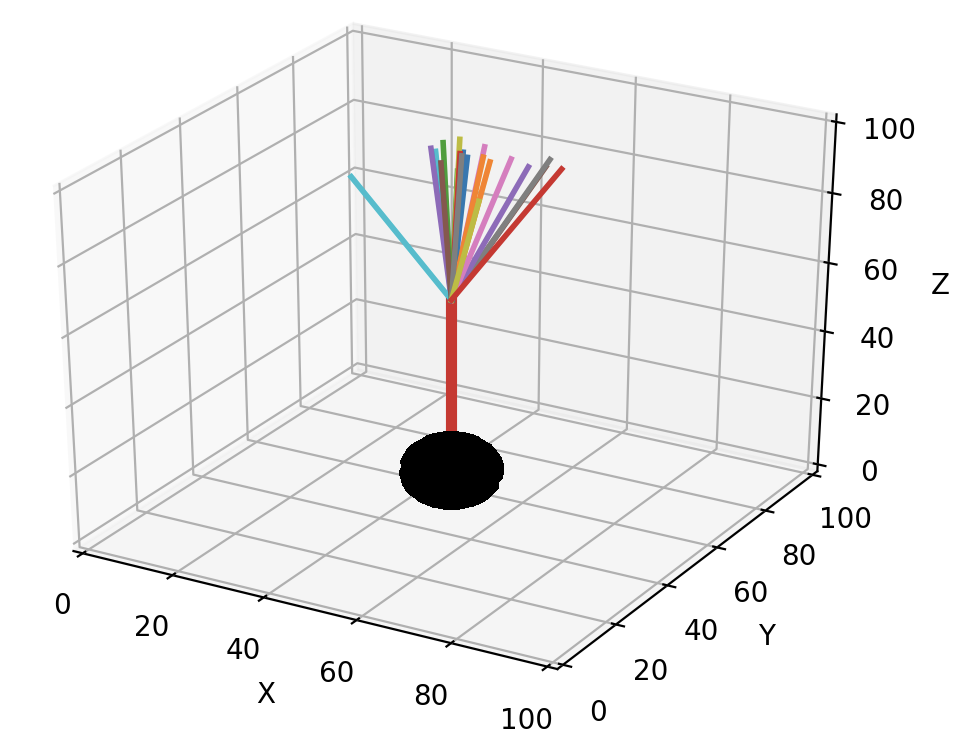
As implicit in the name, unit_heading_sample returns a vector that is 1 µm long. Usually one will want to make longer fronts. In general, fronts should be longer than their radius to avoid spurious collision errors. A simple multiplication of new_dir achieves this goal:
def manage_front(self,constellation):
...
new_dir = self.unit_heading_sample(width=20)
new_pos = self.end + new_dir * 10. # compute position of child end
In the figures above, the different front lengths were 40 µm.
As mentioned in Fronts, front extension will often be determined by combining several influencing factors. Each of these factors can be represented by a vector and the resulting new direction will be the sum of those vectors. For example, the following code:
pia = 199 # z coordinate of the pia, an attractive boundary
...
def manage_front(self,constellation):
...
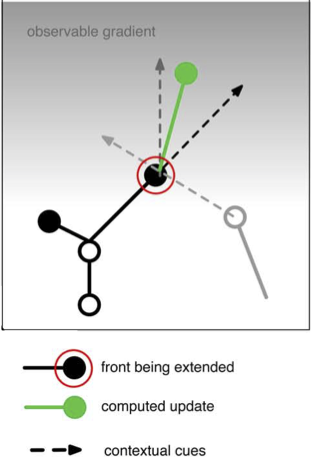
# 1) black arrow: grow in the same direction as self: get the vector direction of self
heading_dir = (self.end - self.orig).norm() # norm returns unit length vector
# 2) light grey arrow: repulsed by another neuron: this requires two steps
# 2a) get a list of positions of all fronts belonging to other nearby neurons, uses the default what="other" option of get_fronts
other_fronts = self.get_fronts(constellation,max_distance=20.)
# 2b) get a direction to the nearest front
if len(other_fronts) > 0: # safe coding!
other = other_fronts[0][0] # closest front
dir_to_other = (other.mid() - self.end).norm() # use mid point on other front
else:
dir_to_other = Point(0.,0.,0.) # no direction
# 3) dark grey arrow: attracted by a gradient: we use the numerically most efficient solution
dir_to_pia = Point(0.,0.,pia).norm() # vertical direction
# now combine by scaling and summing these 3 vectors, repulsion is a subtraction
new_dir = heading_dir * 2.0 - dir_to_other + dir_to_pia * 2.0
new_pos = self.end + new_dir # increment from current positon
...
represents the vector addition in the figure below.
Terminal branching¶
Terminal branching occurs at the growth tips, another form of branching that happens in older parts of the neuron is described in Interstitial branching. Terminal branching is in its simplest form very similar to Front extension but more than one front is created:
def manage_front(self,constellation):
...
points = ... # generate a list of points for new_front.end
rad = self.taper(0.8) # decrease radius
num_branch = 0 # count number of branches
for p in points: # make 2 branches
...
try:
new_front = self.add_child(constellation,p,radius=rad) # make a new front
num_branch += 1 # success
if num_branch == 2: # enough branches made
self.disable(constellation)
return # completed this call
except (CollisionError, GridCompetitionError, InsideParentError, VolumeError):
continue # pick another point, no attempt to correct the error
...
Note that the radius of the new fronts was decreased by 0.8 * self.radius using the taper method. The order of the new fronts is automatically increased by one after each branching event.
Directions for the new branches are chosen with a similar procedure as described in Front extension but using the unit_branching_sample method. unit_branching_sample returns a given number of directions that have an angle relative to the heading of the calling front (if it is cylinder) and a minimal separation between each possible pair. This results a biological branching pattern, the directions are again obtained from normal distributions based on cat spinal cord motor neurons:
points = self.unit_branching_sample(5) # generate more points than needed
An example of default unit_branching_sample:
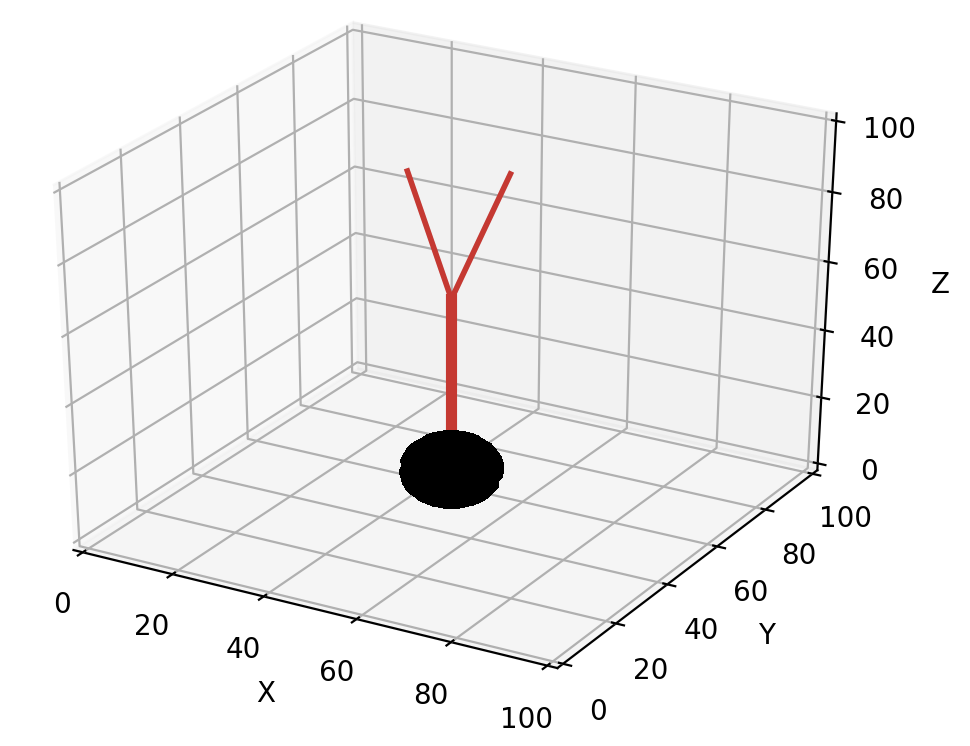
unit_branching_sample can generate a maximum of 20 separated directions. The mean and width of both the branching angle and the separation angle can be changed by the user, see simulator module.
Another requirement is to decide when to branch. The simplest approach is to draw a random number:
def manage_front(self,constellation):
...
if numpy.random.random() < 0.05: # branch
points = self.unit_branching_sample(5)
...
else: # just extend the front
new_dir = self.unit_heading_sample(width=20)
...
This can be made fancier by making the branching probability order-dependent:
def manage_front(self,constellation):
...
if self.order > 5:
bif_prob = 0.03
else: # minimal order for non-somatic front is 1
bif_prob = 0.6 / (self.order * 2.)
if numpy.random.random() < bif_prob: # branch
points = self.unit_branching_sample(5)
...
else: # just extend the front
new_dir = self.unit_heading_sample(width=20)
...
Another approach is to make branching dependent on the environment, for example which cortical layer the front occupies.
Branch termination¶
Growth of a dendritic or axonal branch can be terminated by disabling the front at its tip:
def manage_front(self,constellation):
...
self.disable(constellation) # make inactive and stop growth
return
Obviously a decision is required on when to terminate growth. This is usually done based on random numbers:
def manage_front(self,constellation):
...
if numpy.random.random() < 0.02: # terminate
self.disable(constellation) # make inactive and stop growth
return
Another termination condition can be cumulative distance from the soma path_length:
def manage_front(self,constellation):
...
if self.path_length > 500.: # terminate
self.disable(constellation) # make inactive and stop growth
return
or use self.order, constellation.cycle, etc. An unsolvable CollisionError may also be a reason to terminate growth.
Interstitial branching¶
Interstitial branching is the process where a branch sprouts from a neuron segment that is not a growth cone, this happens more frequently in axons than in dendrites. Simulating interstitial branching is similar to terminal branching but requires careful handling of active and growing status of the parent front. The first step is to not disable the future parent front after it completes its initial front extension:
def manage_front(self,constellation):
...
elif self.swc_type == 4: # apical dendrite: can sprout obliques later
...
try:
new_front = self.add_child(constellation,new_pos,radius=rad) # make a new front and store it
if (self.path_length < 50.): # close to soma, no interstitial growth
self.disable(constellation)
# other new_front are not disabled
return # done for this cycle
except (CollisionError, GridCompetitionError, InsideParentError, VolumeError):
...
In the example code above, taken from the Interstitial Growth notebook, this is done conditionally: only apical dendrite fronts that are some distance from the soma are not disabled.
The interstitial growth itself is handled similarly to front extension but needs to be made a rare event as it should happen for only a few fronts:
def manage_front(self,constellation):
...
elif self.swc_type == 4: # apical dendrite: can sprout obliques later
...
if np.random.random() < 0.0025: # make oblique dendrite
...
try:
new_front = self.add_child(constellation,new_pos,radius=rad,swc_type=8)
self.disable(constellation) # stop interstitial growth
return # done for this cycle
except (CollisionError, GridCompetitionError, InsideParentError, VolumeError):
...
Obviously the code needs to distinguish between front extension and interstitial growth, this can be done by tracking self.num_children:
def manage_front(self,constellation):
...
elif self.swc_type == 4: # apical dendrite: can sprout obliques later
if self.num_children == 0: # initial extension of apical dendrite
# front extension code
...
else:
# interstitial branching code
...
It is easy to generate a growth direction for the oblique dendrite that is close to perpendicular to the apical one by requesting a mean angle of 90 degrees for unit_heading_sample:
rnd_dir = self.unit_heading_sample(mean=90,width=10) # close to perpendicular
new_pos = self.end + rnd_dir * 4.0
It may also be desirable to prevent sprouting of additional oblique dendrites within a given distance of the new one:
def manage_front(self,constellation):
...
elif self.swc_type == 4: # apical dendrite: can sprout obliques later
...
if np.random.random() < 0.0025: # make oblique dendrite
...
rnd_dir = self.unit_heading_sample(mean=90,width=10) # close to perpendicular
new_pos = self.end + rnd_dir * 4.0
try:
new_front = self.add_child(constellation,new_pos,radius=rad,swc_type=8) # make a new front and store it
self.disable(constellation) # stop interstitial growth
# stop interstitial branching within 10 µm distance
neighbors = self.get_neighbors(constellation,10.,branch_stop=True)
for front in neighbors:
if front.is_active():
front.disable(constellation) # stop interstitial growth
return # done for this cycle
except (CollisionError, GridCompetitionError, InsideParentError, VolumeError):
get_neighbors will return a list of fronts that is within a 10 µm path_length distance of self, in both somatopetal and somatofugal directions.
Finally, if oblique growth should occur only at much later developmental stages, it is more efficient to disable the future parent till the cycle in which oblique growth can start:
def manage_front(self,constellation):
...
elif self.swc_type == 4: # apical dendrite: can sprout obliques later
...
try:
new_front = self.add_child(constellation,new_pos,radius=rad) # make a new front and store it
if (self.path_length < 50.): # close to soma, only extension
self.disable(constellation) # no interstitial growth
else:
self.disable(constellation,till_cycle_g=100) # delayed interstitial growth
return # done for this cycle
except (CollisionError, GridCompetitionError, InsideParentError, VolumeError):
...
The till_cycle_g optional parameter disables till the given cycle, at which time the front is made active again and set to growing. Similar code can be used to interrupt any type of growth.